HTE I: Binary treatment
Contents
# pip install -U DoubleML
import random
import pandas as pd
import numpy as np
from scipy.stats import norm
import statsmodels.api as sm
import statsmodels.formula.api as smf
import patsy
from SyncRNG import SyncRNG
import numpy as np
import re
from statsmodels.sandbox.stats.multicomp import multipletests
from scipy import linalg
from itertools import chain
from SyncRNG import SyncRNG
from CTL.causal_tree_learn import CausalTree
from sklearn.model_selection import train_test_split
import plotnine as p
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
4. HTE I: Binary treatment¶
Source RMD file: link
In the previous chapter, we learned how to estimate the effect of a binary treatment averaged over the entire population. However, the average may obscure important details about how different individuals react to the treatment. In this chapter, we will learn how to estimate the conditional average treatment effect (CATE),
which is a “localized” version of the average treatment effect conditional on a vector of observable characteristics.
It’s often the case that (6.1) is too general to be immediately useful, especially when the observable covariates are high-dimensional. It can be hard to estimate reliably without making strong modeling assumptions, and hard to summarize in a useful manner after estimation. In such situations, we will instead try to estimate treatment effect averages for simpler groups
where \(G_i\) indexes subgroups of interest. Below you’ll learn how to estimate and test hypotheses about pre-defined subgroups, and also how to discover subgroups of interest from the data. In this tutorial, you will learn how to use estimates of (6.1) to suggest relevant subgroups \(G_i\) (and in the next chapters you will find out other uses of (6.1) in policy learning and evaluation).
We’ll continue using the abridged version of the General Social Survey (GSS) (Smith, 2016) dataset that was introduced in the previous chapter. In this dataset, individuals were sent to treatment or control with equal probability, so we are in a randomized setting. However, many of the techniques and code shown below should also work in an observational setting provided that unconfoundedness and overlap are satisfied (these assumptions were defined in the previous chapter).
As with other chapters in this tutorial, the code below should still work by replacing the next snippet of code with a different dataset, provided that you update the key variables treatment, outcome, and covariates below. Also, please make sure to read the comments as they may be subtle differences depending on whether your dataset was created in a randomized or observational setting.
# Read in data
data = pd.read_csv( "https://docs.google.com/uc?id=1kSxrVci_EUcSr_Lg1JKk1l7Xd5I9zfRC&export=download" )
n = data.shape[0]
# Treatment: does the the gov't spend too much on "welfare" (1) or "assistance to the poor" (0)
treatment = "w"
# Outcome: 1 for 'yes', 0 for 'no'
outcome = "y"
# Additional covariates
covariates = ["age", "polviews", "income", "educ", "marital", "sex"]
4.1. Pre-specified hypotheses¶
We will begin by learning how to test pre-specified null hypotheses of the form
That is, that the treatment effect is the same regardless of membership to some group \(G_i\). Importantly, for now we’ll assume that the group \(G_i\) was pre-specified – it was decided before looking at the data.
In a randomized setting, if the both the treatment \(W_i\) and group membership \(G_i\) are binary, we can write
This allows us to write the average effects of \(W_i\) and \(G_i\) on \(Y_i\) as
Rewriting the null hypothesis (4.3) in terms of the decomposition (4.5), we see that it boils down to a test about the coefficient in the interaction: \(\beta_{xw} = 0\). Here’s an example that tests whether the treatment effect is the same for “conservative” (polviews < 4) and “liberal” (polviews \(\geq\) 4) individuals.
# Only valid in randomized settings
# Suppose this his group was defined prior to collecting the data
data["conservative"] = np.multiply(data.polviews < 4, 1) # a binary group
group = 'conservative'
# Recall from last chapter -- this is equivalent to running a t-test
fmla = 'y ~ w*conservative'
ols = smf.ols(fmla, data=data).fit(cov_type='HC2')
# print(ols_1.summary())
hypotheses = 'Intercept=0, w=0, conservative=0, w:conservative=0'
t_test = ols.t_test(hypotheses)
print(t_test.summary(xname=list(ols.summary2().tables[1].index)))
Test for Constraints
==================================================================================
coef std err z P>|z| [0.025 0.975]
----------------------------------------------------------------------------------
Intercept 0.4836 0.005 95.127 0.000 0.474 0.494
w -0.3789 0.006 -64.657 0.000 -0.390 -0.367
conservative -0.1590 0.009 -17.195 0.000 -0.177 -0.141
w:conservative 0.1160 0.010 11.185 0.000 0.096 0.136
==================================================================================
Sometimes there are many subgroups, leading to multiple hypotheses such as
In that case, we need to correct for the fact that we are testing for multiple hypotheses, or we will end up with many false positives. The Bonferroni correction (wiki) is a common method for dealing with multiple hypothesis testing, though it is often too conservative to be useful. It is available via the function p.adjust from base R. The next snippet of code tests whether the treatment effect at each level of polviews is different from the treatment effect from polviews equals one.
# Only valid in randomized setting.
# Example: these groups must be defined prior to collecting the data.
group = 'polviews'
# Linear regression.
fmla = 'y ~ w*C(polviews)'
ols = smf.ols(fmla, data=data).fit(cov_type='HC2')
# Retrieve the interaction coefficients
ols_1 = ols.summary2().tables[1].reset_index()
interact = ols_1.loc[ols_1["index"].str.contains("w:")]
hypothesis_1 = []
for i in list(interact['index']):
hypothesis_1.append(i+str('=0'))
hypotheses = hypothesis_1
t_test = ols.t_test(hypotheses)
# print(t_test.summary(xname=list(interact['index'])))
# Retrieve unadjusted p-values and
unadj_p_value = list(interact["P>|z|"])
p_adjusted = list(multipletests(unadj_p_value, alpha=0.05, method='bonferroni')[1])
pd.DataFrame(zip(interact["Coef."], interact["Std.Err."], unadj_p_value, p_adjusted),
columns =['Estimate', 'Std.Err.', 'unadj_p_value', 'adj_p_value'],
index = list(interact["index"]))
| Estimate | Std.Err. | unadj_p_value | adj_p_value | |
|---|---|---|---|---|
| w:C(polviews)[T.2] | -0.024242 | 0.027337 | 3.751982e-01 | 1.000000e+00 |
| w:C(polviews)[T.3] | -0.059623 | 0.027357 | 2.929996e-02 | 1.757997e-01 |
| w:C(polviews)[T.4] | -0.134614 | 0.025340 | 1.082607e-07 | 6.495642e-07 |
| w:C(polviews)[T.5] | -0.164915 | 0.027135 | 1.220112e-09 | 7.320672e-09 |
| w:C(polviews)[T.6] | -0.180079 | 0.027514 | 5.952137e-11 | 3.571282e-10 |
| w:C(polviews)[T.7] | -0.186184 | 0.038706 | 1.507202e-06 | 9.043211e-06 |
# Define a function which turn a list or vector-like object into a proper two
# dimensional column vector
def cvec(a):
""" Turn a list or vector-like object into a proper column vector
Input
a: List or vector-like object, has to be a potential input for np.array()
Output
vec: two dimensional NumPy array, with the first dimension weakly greater
than the second (resulting in a column vector for a vector-like input)
"""
# Conver input into a two dimensional NumPy array
vec = np.array(a, ndmin=2)
# Check whether the second dimension is strictly greater than the first
# (remembering Python's zero indexing)
if vec.shape[0] < vec.shape[1]:
# If so, transpose the input vector
vec = vec.T
# Return the column vector
return vec
def get_cov(X, e, add_intercept=True, homoskedastic=False):
""" Calculates OLS variance estimator based on X and residuals
Inputs
X: n by k matrix, RHS variables
e: n by 1 vector or vector-like, residuals from an OLS regression
add_intercept: Boolean, if True, adds an intercept as the first column of X
(and increases k by one)
Outputs
V_hat: k by k NumPy array, estimated covariance matrix
"""
# Get the number of observations n and parameters k
n, k = X.shape
# Check whether an intercept needs to be added
if add_intercept:
# If so, add the intercept
X = np.concatenate([np.ones(shape=(n,1)), X], axis=1)
# Don't forget to increase k
k = k + 1
# Make sure the residuals are a proper column vector
e = cvec(e)
# Calculate X'X
XX = X.T @ X
# Calculate its inverse
XXinv = linalg.inv(XX)
# Check whether to use homoskedastic errors
if homoskedastic:
# If so, calculate the homoskedastic variance estimator
V_hat = (1 / (n-k)) * XXinv * (e.T @ e)
else:
# Otherwise, calculate an intermediate object
S = (e @ np.ones(shape=(1,k))) * X
# Then, get the HC0 sandwich estimator
V_hat = (n / (n-k)) * XXinv @ (S.transpose() @ S) @ XXinv
# Return the result
return V_hat
# Auxiliary function to computes adjusted p-values
# following the Romano-Wolf method.
# For a reference, see http://ftp.iza.org/dp12845.pdf page 8
# t.orig: vector of t-statistics from original model
# t.boot: matrix of t-statistics from bootstrapped models
def romano_wolf_correction(t_orig, t_boot):
abs_t_orig = np.absolute(t_orig)
abs_t_boot = np.absolute(t_boot)
abs_t_sorted = sorted(abs_t_orig, key = float, reverse=True)
max_order = (-np.array(abs_t_orig)).argsort()
rev_order = np.argsort(max_order)
M = t_boot.shape[0]
S = t_boot.shape[1]
p_adj = list(np.repeat(0, S))
p_adj[0] = np.mean(pd.DataFrame(abs_t_boot).apply(np.max, axis=1) > abs_t_sorted[0])
for s in range(1,S):
cur_index = max_order[s:S]
p_init = np.mean(
pd.DataFrame(abs_t_boot).T.iloc[cur_index].T.apply(np.max, axis=1) > abs_t_sorted[s])
p_adj[s] = np.max(p_init, p_adj[s])
aux = []
for i in rev_order:
aux.append(p_adj[i])
p_adj = aux
return(p_adj)
# Computes adjusted p-values for linear regression (lm) models.
# model: object of lm class (i.e., a linear reg model)
# indices: vector of integers for the coefficients that will be tested
# cov.type: type of standard error (to be passed to sandwich::vcovHC)
# num.boot: number of null bootstrap samples. Increase to stabilize across runs.
# Note: results are probabilitistic and may change slightly at every run.
#
# Adapted from the p_adjust from from the hdm package, written by Philipp Bach.
# https://github.com/PhilippBach/hdm_prev/blob/master/R/p_adjust.R
def summary_rw_lm(model, indices='', cov_type="HC2", num_boot=10000):
SyncRNG(seed = 123456)
# OLS without correction
# Grab the original t values.
ols = smf.ols(fmla, data=data).fit()
ols = ols.summary2().tables[1].reset_index()
summary = ols[ols['index'].isin(list(indices["index"]))]
t_orig = summary['t']
# Null resampling.
# This is a trick to speed up bootstrapping linear models.
# Here, we don't really need to re-fit linear regressions, which would be a bit slow.
# We know that betahat ~ N(beta, Sigma), and we have an estimate Sigmahat.
# So we can approximate "null t-values" by
# - Draw beta.boot ~ N(0, Sigma-hat) --- note the 0 here, this is what makes it a *null* t-value.
# - Compute t.boot = beta.boot / sqrt(diag(Sigma.hat))
ols = smf.ols(fmla, data=data).fit(cov_type = cov_type)
ols_exog = smf.ols(fmla, data=data).exog
ols_res = smf.ols(fmla, data=data).fit().resid
Sigma_hat = get_cov(ols_exog, ols_res, add_intercept=False)
se_orig = pd.Series(np.sqrt(Sigma_hat.diagonal()))
num_coef = len(se_orig)
beta_boot = pd.DataFrame(
np.random.multivariate_normal(
mean=np.repeat(0, num_coef), cov=Sigma_hat, size=num_boot))
t_boot = np.array(beta_boot.apply(lambda row: row / se_orig, axis=1))
t_boot = t_boot.T[(len(ols_1)-len(t_orig)):len(ols_1)].T
p_adj = romano_wolf_correction(t_orig, t_boot)
result = summary[['index','Coef.','Std.Err.','P>|t|']]
result.rename(columns={'P>|t|': 'Orig.p-value'}, inplace=True)
result['Adj. p-value'] = p_adj
return(result)
# This linear regression is only valid in a randomized setting.
fmla = 'y ~ w*C(polviews)'
ols = smf.ols(fmla, data=data).fit()
ols = ols.summary2().tables[1].reset_index()
interact = ols.loc[ols_1["index"].str.contains("w:")]
# Applying the romano-wolf correction.
summary_rw_lm(ols, indices=interact)
C:\Users\Roberto\AppData\Local\Temp\ipykernel_5896\909325009.py:48: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
result.rename(columns={'P>|t|': 'Orig.p-value'}, inplace=True)
C:\Users\Roberto\AppData\Local\Temp\ipykernel_5896\909325009.py:49: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
result['Adj. p-value'] = p_adj
| index | Coef. | Std.Err. | Orig.p-value | Adj. p-value | |
|---|---|---|---|---|---|
| 8 | w:C(polviews)[T.2] | -0.024242 | 0.030348 | 4.244098e-01 | 0.4267 |
| 9 | w:C(polviews)[T.3] | -0.059623 | 0.030150 | 4.798896e-02 | 0.0721 |
| 10 | w:C(polviews)[T.4] | -0.134614 | 0.028228 | 1.862258e-06 | 0.0000 |
| 11 | w:C(polviews)[T.5] | -0.164915 | 0.029575 | 2.481093e-08 | 0.0000 |
| 12 | w:C(polviews)[T.6] | -0.180079 | 0.029660 | 1.284150e-09 | 0.0000 |
| 13 | w:C(polviews)[T.7] | -0.186184 | 0.037904 | 9.063658e-07 | 0.0000 |
The Bonferroni and Romano-Wolf methods control the familywise error rate (FWER), which is the (asymptotic) probability of rejecting even one true null hypothesis. In other words, for a significance level \(\alpha\), they guarantee that with probability \(1 - \alpha\) we will make zero false discoveries. However, when the number of hypotheses being tested is very large, this criterion may be so stringent that it prevents us from being able to detect real effects. Instead, there exist alternative procedures that control the (asymptotic) false discovery rate (FDR), defined as the expected proportion of true null hypotheses rejected among all hypotheses rejected. One such procedure is the Benjamini-Hochberg procedure, which is available in base R via p.adjust(..., method="BH"). For what remains we’ll stick with FWER control, but keep in mind that FDR control can also useful in exploratory analyses or settings in which there’s a very large number of hypotheses under consideration.
As in the previous chapter, when working with observational data under unconfoundedness and overlap, one can use AIPW scores \(\hat{\Gamma}_i\) in place of the raw outcomes \(Y_i\). In the next snippet, we construct AIPW scores using the causal_forest function from the grf package.
4.2. Data-driven hypotheses¶
Pre-specifying hypotheses prior to looking at the data is in general good practice to avoid “p-hacking” (e.g., slicing the data into different subgroups until a significant result is found). However, valid tests can also be attained if by sample splitting: we can use a subset of the sample to find promising subgroups, then test hypotheses about these subgroups in the remaining sample. This kind of sample splitting for hypothesis testing is called honesty.
4.2.1. Via causal trees¶
Causal trees (Athey and Imbens)(PNAS, 2016) are an intuitive algorithm that is available in the randomized setting to discover subgroups with different treatment effects.
At a high level, the idea is to divide the sample into three subsets (not necessarily of equal size). The splitting subset is used to fit a decision tree whose objective is modified to maximize heterogeneity in treatment effect estimates across leaves. The estimation subset is then used to produce a valid estimate of the treatment effect at each leaf of the fitted tree. Finally, a test subset can be used to validate the tree estimates.
The next snippet uses honest.causalTree function from the causalTree package. For more details, see the causalTree documentation.
X = data[['age','polviews', 'income','educ','marital','sex']]
y = data['y']
treatment = data['w']
columns = X.columns # get varaible's names in a list
# From DataFrame to array
X = X.values
y = y.values
treatment = treatment.values
# CL-honest
cthl = CausalTree(honest=True, min_size=1, split_size=0.33)
cthl.fit(X, y, treatment)
cthl.prune()
# plot
cthl.plot_tree(features=columns, filename="bin_tree_honest_1", show_effect=True, alpha = 0)
C:\Users\Roberto\anaconda3\lib\site-packages\numpy\core\fromnumeric.py:3723: RuntimeWarning: Degrees of freedom <= 0 for slice
C:\Users\Roberto\anaconda3\lib\site-packages\numpy\core\_methods.py:254: RuntimeWarning: invalid value encountered in double_scalars
# get train and test sample
train_x, val_x, train_y, val_y, train_t, val_t = train_test_split(X, y, treatment, random_state=724, shuffle=True,
test_size=0.33)
# get honest/estimation portion
train_x, est_x, train_y, est_y, train_t, est_t = train_test_split(train_x, train_y, train_t, shuffle=True,
random_state=724, test_size=0.5)
cthl_predict = cthl.predict(est_x) # Predict tau
num_leaves = len(np.unique(cthl_predict)) # number of leaves
labels = [i for i in range(1,len(np.unique(cthl_predict)) + 1 ) ] # label by each leaf
# Prediction grouped by each leaf
predict = pd.DataFrame({"predict": cthl_predict})
predict['leaves'] = pd.Categorical(predict.predict)
predict['leaves'] = predict['leaves'].cat.rename_categories(labels)
predict
| predict | leaves | |
|---|---|---|
| 0 | -0.369901 | 7 |
| 1 | -0.369901 | 7 |
| 2 | -0.369901 | 7 |
| 3 | -0.369901 | 7 |
| 4 | -0.369901 | 7 |
| ... | ... | ... |
| 9594 | -0.406130 | 5 |
| 9595 | -0.433803 | 3 |
| 9596 | -0.250101 | 10 |
| 9597 | -0.369901 | 7 |
| 9598 | -0.369901 | 7 |
9599 rows × 2 columns
4.2.1.1. Via grf - Causal Forest¶
#!pip install econml
import econml
# Main imports
from econml.orf import DMLOrthoForest, DROrthoForest
from econml.dml import CausalForestDML
from econml.sklearn_extensions.linear_model import WeightedLassoCVWrapper, WeightedLasso, WeightedLassoCV
from sklearn.linear_model import MultiTaskLassoCV
# Helper imports
import numpy as np
from itertools import product
from sklearn.linear_model import Lasso, LassoCV, LogisticRegression, LogisticRegressionCV
import matplotlib.pyplot as plt
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from econml.grf import RegressionForest
%matplotlib inline
import patsy
import seaborn as sns
# Preparing data to fit a causal forest
fmla = '0+age+polviews+income+educ+marital+sex'
desc = patsy.ModelDesc.from_formula(fmla)
desc.describe()
matrix = patsy.dmatrix(fmla, data, return_type = "dataframe")
T = data.loc[ : ,"w"]
Y = data.loc[ : ,"y"]
X = matrix
W = None
# Estimate a causal forest.
est2 = CausalForestDML(model_t=RegressionForest(),
model_y=RegressionForest(),
n_estimators=200, min_samples_leaf=5,
max_depth=50,
verbose=0, random_state=123)
est2.tune(Y, T, X=X, W=W)
est2.fit(Y, T, X=X, W=W)
<econml.dml.causal_forest.CausalForestDML at 0x1fb09195490>
# Get residuals and propensity
residuals = est2.fit(Y, T, X=X, W=W, cache_values=True).residuals_
T_res = residuals[1]
e_hat = T - T_res
# T = beta_hat*X + e , beta_hat*X = T_hat = T-e
Prop = pd.DataFrame({"p_score":e_hat, "Treatment":T})
# Propensity score
# Do not use this for assessing heterogeneity. See text above.
sns.histplot(data=Prop, x="p_score", hue="Treatment", bins=40, alpha = 0.4)
<AxesSubplot:xlabel='p_score', ylabel='Count'>
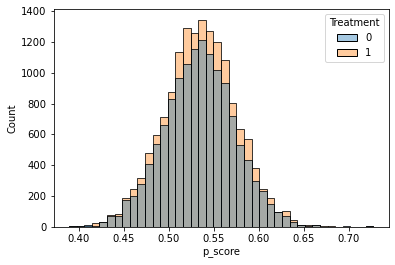
Having fit a non-parametric method such as a causal forest, a researcher may (incorrectly) start by looking at the distribution of its predictions of the treatment effect. One might be tempted to think: “if the histogram is concentrated at a point, then there is no heterogeneity; if the histogram is spread out, then our estimator has found interesting heterogeneity.” However, this may be false.
tau_hat = est2.effect(X=X) # tau(X) estimates
# Do not use this for assessing heterogeneity. See text above.
sns.displot( tau_hat, stat="density", bins = 10)
plt.title("CATE estimates")
Text(0.5, 1.0, 'CATE estimates')
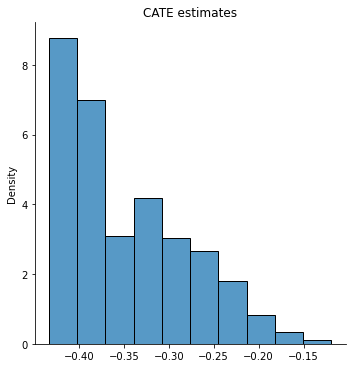
If the histogram is concentrated at a point, we may simply be underpowered: our method was not able to detect any heterogeneity, but maybe it would detect it if we had more data. If the histogram is spread out, we may be overfitting: our model is producing very noisy estimates \(\widehat{\tau}(x)\), but in fact the true \(\tau(x)\) can be much smoother as a function of \(x\).
The grf package also produces a measure of variable importance that indicates how often a variable was used in a tree split. Again, much like the histogram above, this can be a rough diagnostic, but it should not be interpreted as indicating that, for example, variable with low importance is not related to heterogeneity. The reasoning is the same as the one presented in the causal trees section: if two covariates are highly correlated, the trees might split on one covariate but not the other, even though both (or maybe neither) are relevant in the true data-generating process.
est2.feature_importances()
array([0.05183671, 0.49923342, 0.37411192, 0.0567607 , 0.01369339,
0.00436387])
importance = pd.DataFrame({"covariaties" : list(X.columns), "values" : est2.feature_importances()})
importance.sort_values('values', ascending = False)
| covariaties | values | |
|---|---|---|
| 1 | polviews | 0.499233 |
| 2 | income | 0.374112 |
| 3 | educ | 0.056761 |
| 0 | age | 0.051837 |
| 4 | marital | 0.013693 |
| 5 | sex | 0.004364 |
4.2.1.2. Data-driven subgroups¶
Just as with causal trees, we can use causal forests to divide our observations into subgroups. In place of leaves, we’ll rank observation into (say) quintiles according to their estimated CATE prediction; see, e.g., Chernozhukov, Demirer, Duflo, Fernández-Val (2020) for similar ideas.
There’s a subtle but important point that needs to be addressed here. As we have mentioned before, when predicting the conditional average treatment effect \(\tau(X_i)\) for observation \(i\) we should in general avoid using a model that was fitted using observation \(i\). This sort of sample splitting (which we called honesty above) is one of the required ingredients to get unbiased estimates of the CATE using the methods described here. However, when ranking estimates of two observations \(i\) and \(j\), we need something a little stronger: we must ensure that the model was not fit using either \(i\) or \(j\)’s data.
One way of overcoming this obstacle is simple. First, divide the data into \(K\) folds (subsets). Then, cycle through the folds, fitting a CATE model on \(K-1\) folds. Next, for each held-out fold, separately rank the unseen observations into \(Q\) groups based on their prediction (i.e., if \(Q=5\), then we rank observations by estimated CATE into “top quintile”, “second quintile”, and so on). After concatenating the independent rankings together, we can study the differences in observations in each rank-group.
This gist computes the above for grf, and it should not be hard to modify it so as to replace forests by any other non-parametric method. However, for grf specifically, there’s a small trick that allows us to obtain a valid ranking: we can pass a vector of fold indices to the argument clusters and rank observations within each fold. This works because estimates for each fold (“cluster”) trees are computed using trees that were not fit using observations from that fold. Here’s how to do it.
def cluster_causal_forest(Y,T, X, W, cluster):
base = pd.concat([Y,T], axis = 1)
for i in range(cluster):
index=range(X.shape[0])
a = np.array_split(np.array(index),cluster)[i] ## split index
Y = base.drop(base.iloc[list(a),:].index).iloc[:,0]
T = base.drop(base.iloc[list(a),:].index).iloc[:,1]
XX = X.drop(X.iloc[list(a),:].index)
causal = CausalForestDML(model_t=RegressionForest(),
model_y=RegressionForest(),
n_estimators=200, min_samples_leaf=5,
max_depth=50,
verbose=0, random_state=123)
causal.fit(Y, T, X=XX, W=W)
tau_hat = causal.effect(X=X.iloc[list(a),:]) # tau(X) estimates using validation data
vector = i*np.ones( len(list(a)) ) + 1
globals()[f'data_{i}'] = pd.DataFrame({"tau_hat":tau_hat, "Cluster":vector})
tau_predict = data_0.copy()
for k in range(1,10):
tau_predict = tau_predict.append(globals()[f'data_{k}'] , ignore_index=True)
return tau_predict
The next snippet computes the average treatment effect within each group defined above, i.e., \(\mathbf{E}[Y_{i}(0)|G_{i} = g]\). This can done in two ways. First, by computing a simple difference-in-means estimate of the ATE based on observations within each group. This is valid only in randomized settings.
# Valid randomized data and observational data with unconfoundedness+overlap.
# Note: read the comments below carefully.
# In randomized settings, do not estimate forest.e and e.hat; use known assignment probs.
# Prepare dataset
fmla = '0+age+polviews+income+educ+marital+sex'
desc = patsy.ModelDesc.from_formula(fmla)
matrix = patsy.dmatrix(fmla, data, return_type = "dataframe")
T = data.loc[ : ,"w"]
Y = data.loc[ : ,"y"]
X = matrix
W = None
# Number of rankings that the predictions will be ranking on
# (e.g., 2 for above/below median estimated CATE, 5 for estimated CATE quintiles, etc.)
num_rankings = 5
# Prepare for data.splitting
# Assign a fold number to each observation.
# The argument 'clusters' in the next step will mimick K-fold cross-fitting.
num_folds = 10
# Estimate a causal forest.
tau_hat_cluster = cluster_causal_forest(Y,T, X, W, cluster = num_folds)
data['ranking'] = np.nan
for i in range(1,num_folds+1):
split = tau_hat_cluster.Cluster == i
index = tau_hat_cluster[split].index
tau_quantile = np.quantile(tau_hat_cluster[split].iloc[:,0], list(np.arange(0,1.1,0.2)))
labels =[i for i in range(1,6)]
data.loc[index , ["ranking"]] = pd.cut(tau_hat_cluster[split]["tau_hat"],
tau_quantile , right=False, labels=labels)
The frame.append method is deprecated and will be removed from pandas in a future version. Use pandas.concat instead.
# Valid only in randomized settings.
# Average difference-in-means within each ranking
# Formula y ~ 0 + ranking + ranking:w
fmla = 'y ~ 0 + C(ranking) + w:C(ranking)'
ols = smf.ols(fmla, data=data).fit(cov_type='HC2')
# Retrieve the interaction coefficients
ols_1 = ols.summary2().tables[1].reset_index()
ols_ate = ols_1.loc[ols_1["index"].str.contains("w:")].iloc[:,1:3]
ols_ate['method'] = "ols"
ols_ate['ranking'] = [f'Q{j}' for j in range(1,6)]
order = [2,3,0,1] # setting column's order
ols_ate = ols_ate[[ols_ate.columns[i] for i in order]]
ols_ate.rename({'Coef.': 'Estimate', 'Std.Err.': 'se'}, axis=1, inplace = True)
ols_ate
| method | ranking | Estimate | se | |
|---|---|---|---|---|
| 5 | ols | Q1 | -0.435064 | 0.011338 |
| 6 | ols | Q2 | -0.397305 | 0.011151 |
| 7 | ols | Q3 | -0.361381 | 0.010971 |
| 8 | ols | Q4 | -0.320245 | 0.010623 |
| 9 | ols | Q5 | -0.223207 | 0.009822 |
Another option is to average the AIPW scores within each group. This valid for both randomized settings and observational settings with unconfoundedness and overlap. Moreover, AIPW-based estimators should produce estimates with tighter confidence intervals in large samples.
# Computing AIPW scores.
tau_hat = est2.effect(X=X) #E[Y|X]
residuals = est2.fit(Y, T, X=X, W=W, cache_values=True).residuals_
T_res = residuals[1]
e_hat = T - T_res # P[W=1|X]
Y_res = residuals[0]
m_hat = Y - Y_res # E[Y|X]
# Estimating mu.hat(X, 1) and mu.hat(X, 0) for obs in held-out sample
# Note: to understand this, read equations 6-8 in this vignette:
# https://grf-labs.github.io/grf/articles/muhats.html
mu_hat0 = m_hat - e_hat * tau_hat # E[Y|X,W=0] = E[Y|X] - e(X)*tau(X)
mu_hat1 = m_hat + (1 - e_hat) * tau_hat # E[Y|X,W=1] = E[Y|X] + (1 - e(X))*tau(X)
# AIPW scores
data['aipw_scores'] = tau_hat + T / e_hat * (Y - mu_hat1) - (1 - T) / (1 - e_hat) * (Y - mu_hat0)
fmla = 'aipw_scores ~ 0 + C(ranking)'
ols = smf.ols(fmla,data).fit(cov_type='HC2')
# Retrieve the interaction coefficients
ols = ols.summary2().tables[1].reset_index()
forest_ate = ols.loc[ols["index"].str.contains("ranking")].iloc[:,1:3]
forest_ate['method'] = "apiw"
forest_ate['ranking'] = [f'Q{j}' for j in range(1,6)]
order = [2,3,0,1] # setting column's order
forest_ate = forest_ate[[forest_ate.columns[i] for i in order]]
forest_ate.rename({'Coef.': 'Estimate', 'Std.Err.': 'se'}, axis=1, inplace = True)
forest_ate
| method | ranking | Estimate | se | |
|---|---|---|---|---|
| 0 | apiw | Q1 | -0.430011 | 0.011363 |
| 1 | apiw | Q2 | -0.395391 | 0.011210 |
| 2 | apiw | Q3 | -0.362498 | 0.011041 |
| 3 | apiw | Q4 | -0.322003 | 0.010707 |
| 4 | apiw | Q5 | -0.222019 | 0.009806 |
The code below plots the estimates.
# Concatenate the two results.
res = pd.concat([forest_ate, ols_ate])
# Plotting the point estimate of average treatment effect
# and 95% confidence intervals around it.
# Define figure, axes, and plot
fig, ax = plt.subplots(figsize=(10, 6))
ax.scatter(x=res['ranking'].unique(),
marker='o', s=40,
y=res[res.method == "ols"]['Estimate'], color = "blue")
ax.scatter(x=np.arange(0.1,5,1),
marker='o', s=40,
y=res[res.method == "apiw"]['Estimate'], color = "red")
eb1 = plt.errorbar(x=res['ranking'].unique(), y=res[res.method == "ols"]['Estimate'],
yerr= 2*res[res.method == "ols"]['se'],
color = 'darkblue', ls='', capsize = 4)
eb1[-1][0].set_linestyle('--')
eb2 = plt.errorbar(x=np.arange(0.1,5,1), y=res[res.method == "apiw"]['Estimate'],
yerr= 2*res[res.method == "apiw"]['se'],
color = 'darkred', ls='', capsize = 4)
eb2[-1][0].set_linestyle('--')
# Set title & labels
plt.title('Average CATE within each ranking (as defined by predicted CATE)',fontsize=15)
# add legend
red_patch = mpatches.Patch(color='red', label='apiw')
blue_patch = mpatches.Patch(color='blue', label='ols')
plt.legend(title = "Method", handles=[blue_patch,red_patch], bbox_to_anchor=(1.15, 0.7))
plt.show()
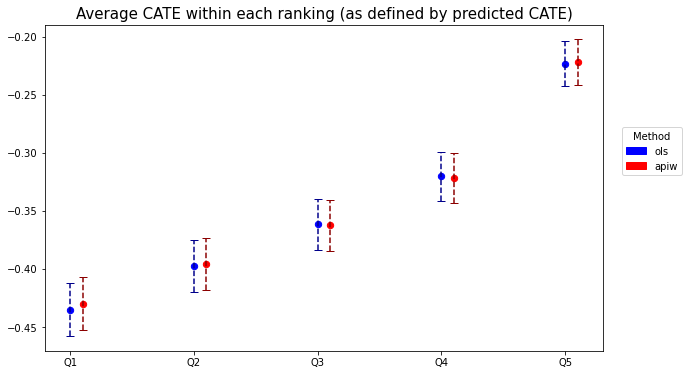
When there isn’t much detectable heterogeneity, the plot above can end up being non-monotonic. This can mean that the number of observations is too small for us to be able to detect subgroups with relevant differences in treatment effect.
As an exercise, try running the previous two snippets on few data points (e.g., the first thousand observations only). You will likely see the "non-monotonicity" phenomenon just described.Next, as we did for leaves in a causal tree, we can test e.g., if the prediction for groups 2, 3, etc. are larger than the one in the first group. Here’s how to do it based on a difference-in-means estimator. Note the Romano-Wolf multiple-hypothesis testing correction.
# Valid in randomized settings only.
# y ~ ranking + w + ranking:w
fmla = 'y ~ C(ranking) + w + w:C(ranking)'
ols = smf.ols(fmla, data=data).fit()
# Retrieve the interaction coefficients
ols_1 = ols.summary2().tables[1].reset_index()
interact = ols_1.loc[ols_1["index"].str.contains("w:")]
ols_ate = summary_rw_lm(ols, indices=interact)
ols_ate['ranking'] = [f'Rank {j} - Rank 1' for j in range(2,6)]
order = [5,1,2,3,4] # setting column's order
ols_ate = ols_ate[[ols_ate.columns[i] for i in order]]
ols_ate.rename({'Coef.': 'Estimate', 'Std.Err.': 'se', 'ranking': 'Comparative'}, axis=1, inplace = True)
ols_ate
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
| Comparative | Estimate | se | Orig.p-value | Adj. p-value | |
|---|---|---|---|---|---|
| 6 | Rank 2 - Rank 1 | 0.037759 | 0.014777 | 1.061615e-02 | 0.0101 |
| 7 | Rank 3 - Rank 1 | 0.073683 | 0.014772 | 6.136161e-07 | 0.0000 |
| 8 | Rank 4 - Rank 1 | 0.114819 | 0.014771 | 7.909513e-15 | 0.0000 |
| 9 | Rank 5 - Rank 1 | 0.211857 | 0.014779 | 1.919290e-46 | 0.0000 |
Here’s how to do it for AIPW-based estimates, again with Romano-Wolf correction for multiple hypothesis testing.
# Valid in randomized and observational settings with unconfoundedness+overlap.
# Using AIPW scores computed above
fmla = 'aipw_scores ~ 1 + C(ranking)'
ols = smf.ols(fmla, data=data).fit()
# Retrieve the interaction coefficients
ols_1 = ols.summary2().tables[1].reset_index()
interact = ols_1.loc[1:num_rankings +1 ,:]
forest_ate = summary_rw_lm(ols, indices=interact)
forest_ate['ranking'] = [f'Rank {j} - Rank 1' for j in range(2,6)]
order = [5,1,2,3,4] # setting column's order
forest_ate = forest_ate[[forest_ate.columns[i] for i in order]]
forest_ate.rename({'Coef.': 'Estimate', 'Std.Err.': 'se', 'ranking': 'Comparative'}, axis=1, inplace = True)
forest_ate
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
| Comparative | Estimate | se | Orig.p-value | Adj. p-value | |
|---|---|---|---|---|---|
| 1 | Rank 2 - Rank 1 | 0.034621 | 0.015330 | 2.392637e-02 | 0.0229 |
| 2 | Rank 3 - Rank 1 | 0.067514 | 0.015328 | 1.063206e-05 | 0.0000 |
| 3 | Rank 4 - Rank 1 | 0.108008 | 0.015330 | 1.891189e-12 | 0.0000 |
| 4 | Rank 5 - Rank 1 | 0.207993 | 0.015334 | 8.835049e-42 | 0.0000 |
Finally, we can also check if different groups have different average covariate levels across rankings.
df = pd.DataFrame()
for var_name in covariates:
# Looping over covariate names
# Compute average covariate value per ranking (with correct standard errors)
form2 = var_name + " ~ " + "0" + "+" + "C(ranking)"
ols = smf.ols(formula=form2, data=data).fit(cov_type = 'HC2').summary2().tables[1].iloc[:, 0:2]
# Retrieve results
toget_index = ols["Coef."]
index = toget_index.index
cova1 = pd.Series(np.repeat(var_name,num_rankings), index = index, name = "covariate")
avg = pd.Series(ols["Coef."], name="avg")
stderr = pd.Series(ols["Std.Err."], name = "stderr")
ranking = pd.Series(np.arange(1,num_rankings+1), index = index, name = "ranking")
scaling = pd.Series(norm.cdf((avg - np.mean(avg))/np.std(avg)), index = index, name = "scaling")
data2 = pd.DataFrame(data=X, columns= covariates)
variation1= np.std(avg) / np.std(data2[var_name])
variation = pd.Series(np.repeat(variation1, num_rankings), index = index, name = "variation")
labels = pd.Series(round(avg,2).astype('str') + "\n" + "(" + round(stderr, 2).astype('str') + ")", index = index, name = "labels")
# Tally up results
df1 = pd.DataFrame(data = [cova1, avg, stderr, ranking, scaling, variation, labels]).T
df = df.append(df1)
# a small optional trick to ensure heatmap will be in decreasing order of 'variation'
df = df.sort_values(by = ["variation", "covariate"], ascending = False)
df = df.iloc[0:(8*num_rankings), :]
df1 = df.pivot(index = "covariate", columns = "ranking", values = ["scaling"]).astype(float)
labels = df.pivot(index = "covariate", columns = "ranking", values = ["labels"]).to_numpy()
# plot heatmap
ax = plt.subplots(figsize=(10, 10))
ax = sns.heatmap(df1,
annot=labels,
annot_kws={"size": 12, 'color':"k"},
fmt = '',
cmap = "YlGn",
linewidths=0,
xticklabels = ranking)
plt.tick_params( axis='y', labelsize=15, length=0, labelrotation=0)
plt.tick_params( axis='x', labelsize=15, length=0, labelrotation=0)
plt.xlabel("CATE estimate ranking", fontsize= 10)
plt.ylabel("")
ax.set_title("Average covariate values within group (based on CATE estimate ranking)", fontsize=18, fontweight = "bold")
(ax.set_xticklabels(["Q1", "Q2", "Q3", "Q4","Q5"], size=15))
The frame.append method is deprecated and will be removed from pandas in a future version. Use pandas.concat instead.
The frame.append method is deprecated and will be removed from pandas in a future version. Use pandas.concat instead.
The frame.append method is deprecated and will be removed from pandas in a future version. Use pandas.concat instead.
The frame.append method is deprecated and will be removed from pandas in a future version. Use pandas.concat instead.
The frame.append method is deprecated and will be removed from pandas in a future version. Use pandas.concat instead.
The frame.append method is deprecated and will be removed from pandas in a future version. Use pandas.concat instead.
[Text(0.5, 0, 'Q1'),
Text(1.5, 0, 'Q2'),
Text(2.5, 0, 'Q3'),
Text(3.5, 0, 'Q4'),
Text(4.5, 0, 'Q5')]
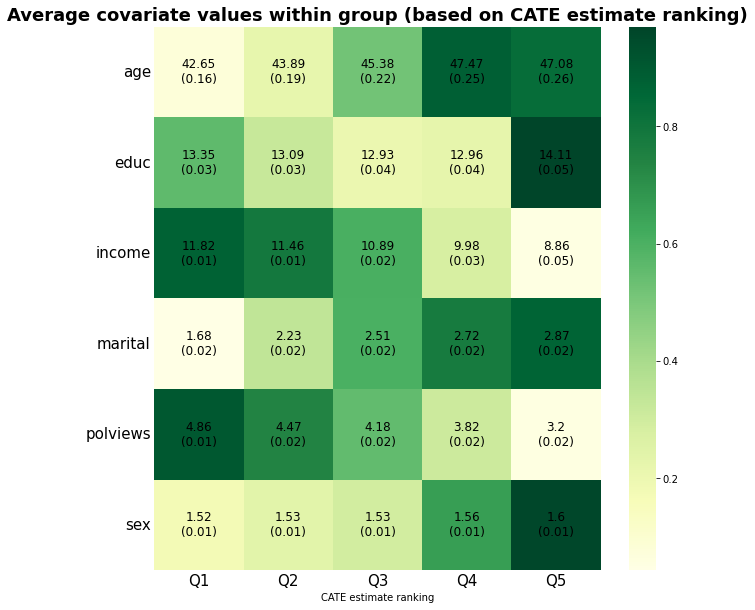
Interpret the heatmap above. What describes the subgroups with strongest and weakest estimated treatment effect? Are there variables that seem to increase or decrease monotonically across rankings?
4.2.1.3. Best linear projection¶
This function provides a doubly robust fit to the linear model \(\widehat{\tau}(X_i) = \beta_0 + A_i'\beta_1\), where \(A_i\) can be a subset of the covariate columns. The coefficients in this regression are suggestive of general trends, but they should not be interpret as partial effects – that would only be true if the true model were really linear in covariates, and that’s an assumption we shouldn’t be willing to make in general
# dataset from best linear proyection
d = X
d['tau_hat'] = tau_hat
smf.ols('m_hat ~ age+polviews+income+educ+marital+sex',d).fit(cov_type = 'HC3').summary2().tables[1]
| Coef. | Std.Err. | z | P>|z| | [0.025 | 0.975] | |
|---|---|---|---|---|---|---|
| Intercept | 0.029668 | 0.002917 | 10.169237 | 2.720309e-24 | 0.023950 | 0.035386 |
| age | -0.000240 | 0.000020 | -12.226411 | 2.246461e-34 | -0.000278 | -0.000201 |
| polviews | 0.038560 | 0.000277 | 139.322948 | 0.000000e+00 | 0.038017 | 0.039102 |
| income | 0.012015 | 0.000146 | 82.440837 | 0.000000e+00 | 0.011729 | 0.012301 |
| educ | -0.001681 | 0.000123 | -13.620749 | 3.014620e-42 | -0.001923 | -0.001439 |
| marital | -0.006065 | 0.000210 | -28.886082 | 1.785792e-183 | -0.006476 | -0.005653 |
| sex | -0.008247 | 0.000630 | -13.094279 | 3.550317e-39 | -0.009482 | -0.007013 |
4.2.1.4. Partial dependence¶
It may also be interesting to examine how our CATE estimates behave when we change a single covariate, while keeping all the other covariates at a some fixed value. In the plot below we evaluate a variable of interest across quantiles, while keeping all other covariates at their median.
It is important to recognize that by keeping some variables at their median we may be evaluating the CATE at \(x\) values in regions where there are few or no data points. Also, it may be the case that varying some particular variable while keeping others fixed may just not be very interesting.
In what follows we’ll again use causal_forest predictions, along with their variance estimates (set estimate.variances=TRUE when predicting to we get estimates of the asymptotic variance of the prediction for each point). Since grf predictions are asymptotically normal, we can construct 95% confidence intervals in the usual manner (i.e., \(\hat{\tau}(x) \pm 1.96\sqrt{\widehat{\text{Var}}(\hat{\tau}(x))}\)).
selected_covariate = "polviews"
other_covariates = ["age", "income", "educ", "marital", "sex"]
# Fitting a forest
# (commented for convenience; no need re-fit if already fitted above)
fmla = '0+age+polviews+income+educ+marital+sex'
# Compute a grid of values appropriate for the selected covariate
grid_size = 7
covariate_grid = np.array_split(data[selected_covariate].sort_values().unique(),grid_size)
# Take median of other covariates
median = data[other_covariates].median(axis = 0).to_numpy().reshape(1,5)
# # Construct a dataset
data_grid = pd.concat([pd.DataFrame(median, columns = other_covariates)]*grid_size)
data_grid[selected_covariate ] = [i[0] for i in covariate_grid]
# Expand the data
X_grid = patsy.dmatrix(fmla, data_grid, return_type = "dataframe")
# Point predictions of the CATE and standard errors
tau_hat = est2.effect(X=X_grid)
tau_hat_ci = est2.effect_interval(X=X_grid)
tau_hat_se = (tau_hat_ci[1]-tau_hat_ci[0])/2
1# Plot predictions for each group and 95% confidence intervals around them.
fig, ax = plt.subplots(figsize=(10, 6))
ax.scatter(x=data_grid[selected_covariate],
marker='o', s=40,
y=tau_hat, color = "blue")
eb1 = plt.errorbar(x=data_grid[selected_covariate], y=tau_hat,
yerr= tau_hat_se,
color = 'darkblue', ls='', capsize = 4)
eb1[-1][0].set_linestyle('--')
ax.set_title(f"Predicted treatment effect varying {selected_covariate} (other variables fixed at median)",
fontsize=11, fontweight = "bold")
(ax.set_xticklabels(range(0,8), size=10))
FixedFormatter should only be used together with FixedLocator
[Text(0.0, 0, '0'),
Text(1.0, 0, '1'),
Text(2.0, 0, '2'),
Text(3.0, 0, '3'),
Text(4.0, 0, '4'),
Text(5.0, 0, '5'),
Text(6.0, 0, '6'),
Text(7.0, 0, '7'),
Text(8.0, 0, '')]
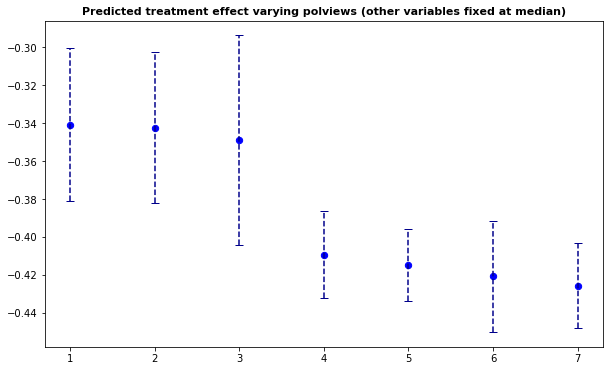
Note that, in this example, we got fairly large confidence intervals. This can be explained in two ways. First, as we mentioned above, there’s a data scarcity problem.
As documented in the original paper on generalized random forests (Athey, Tibshirani and Wager, 2019), the coverage of grf confidence intervals can drop if the signal is too dense or too complex relative to the number of observations. Also, to a point, it’s possible to get tighter confidence intervals by increasing the number of trees; see this short vignette for more information.
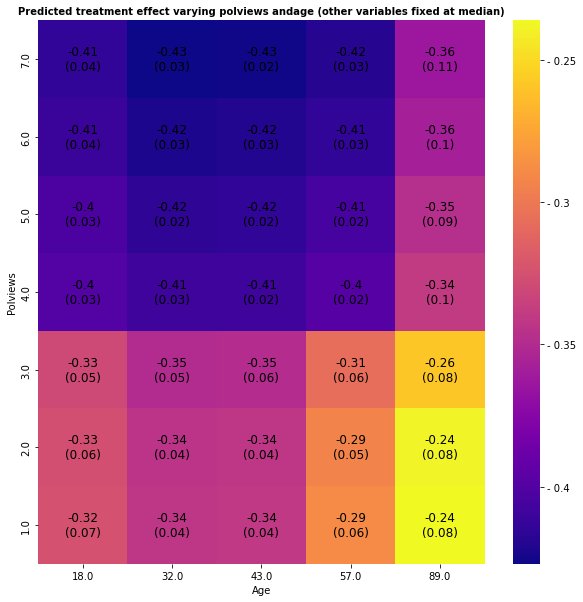
We can vary more than one variable at a time. The next snippet shows predictions and standard errors varying two variables.
selected_covariates = ['polviews','age' ]
other_covariates = ["income", "educ", "marital", "sex"]
# Compute a grid of values appropriate for the selected covariate
# See other options for constructing grids in the snippet above.
x1_grid_size = 7
x2_grid_size = 5
x1_grid = np.array_split(data[selected_covariate].sort_values().unique(),grid_size)
x2_grid = np.quantile(data[selected_covariates[1]], list(np.arange(0,1.01,1/4)))
# Take median of other covariates
median = data[other_covariates].median(axis = 0).to_numpy().reshape(1,4)
# Construct dataset
data_grid = pd.concat([pd.DataFrame(median, columns = other_covariates)]*(x1_grid_size))
data_grid[selected_covariates[0]] = [i[0] for i in covariate_grid]
data_grid = pd.concat([data_grid]*(x2_grid_size))
data_grid[selected_covariates[1]] = list(pd.Series(x2_grid).repeat(7))
# Expand the data
X_grid = patsy.dmatrix(fmla, data_grid, return_type = "dataframe")
# Forest-based point estimates of CATE and standard errors around them
tau_hat = est2.effect(X=X_grid)
tau_hat_ci = est2.effect_interval(X=X_grid)
tau_hat_se = (tau_hat_ci[1]-tau_hat_ci[0])/2
# A vector of labels for plotting below
df = X_grid
df['tau_hat'] = tau_hat
df['tau_hat_se'] = tau_hat_se
# order dataset
df = df.sort_values(by = ["polviews","age"], ascending = [False,True])
labels = np.array(round(df.tau_hat,2).astype('str') + "\n" + "(" + round(df.tau_hat_se, 2).astype('str') + ")")
labels = labels.reshape(7,5) # reshape labels
df = df.pivot("polviews","age","tau_hat") # reshape dataset
df = df.sort_values(by = ["polviews"], ascending = False)
# Plotting
cbar_ticks = [-0.4,-0.35,-0.3,-0.25]
ax = plt.subplots(figsize=(10, 10))
ax = sns.heatmap(df,
annot=labels,
annot_kws={"size": 12, 'color':"k"},
fmt = '',
cmap = "plasma",
linewidths=0,
cbar_kws={"pad":0.05, "ticks": cbar_ticks})
cbar = ax.collections[0].colorbar
cbar.set_ticklabels(['- 0.4', '- 0.35', '- 0.3', '- 0.25'])
plt.xlabel("Age", fontsize= 10)
plt.ylabel("Polviews", fontsize= 10)
ax.set_title(f"Predicted treatment effect varying {selected_covariates[0]} and{selected_covariates[1]} (other variables fixed at median)",
fontsize=10, fontweight = "bold")
Text(0.5, 1.0, 'Predicted treatment effect varying polviews andage (other variables fixed at median)')
4.3. Further reading¶
A readable summary of different method for hypothesis testing correction is laid out in the introduction to Clarke, Romano and Wolf (2009).
Athey and Wager (2019) shows an application of causal forests to heterogeity analysis in a setting with clustering.
